The hidden diversity of widespread predatory amoebae of the order Vampyrellida (Rhizaria)

The order Vampyrellida comprises predatory amoebae forming a large and genetically diverse clade of rhizarian protists. We know that these intricate microbes specifically prey on other eukaryotes, but their actual diversity and ecological roles are still fragmentarily known. We sample freshwater, marine and terrestrial habitats, establish vampyrellid cultures by single cell isolation, study their cell structure by light and electron microscopy, and shed light on their feeding ecology with lab experiments. This is combined with next-generation sequencing techniques to assess the genetic vampyrellid diversity in poorly sampled habitats, and to resolve the evolutionary relationships of the major vampyrellid clades (families). Our aim is to further our phenotypic understanding of these widespread and ecologically interesting microbes, and provide a basis for their identification.
Funded by the German Research Foundation, DFG (Taxon-omics, SPP 1991); 2021-2023
- Methods: Cell isolation, cultivation of protists, light- and electron microscopy, molecular phylogenetics, Illumina short read sequencing, taxonomy
- Relevant publications: Hess et al. 2012, Hess 2017a, Hess 2017b, More et al. 2018
Carbohydrate-active enzymes used in cell wall penetration by unicellular predators and parasitoids of microalgae
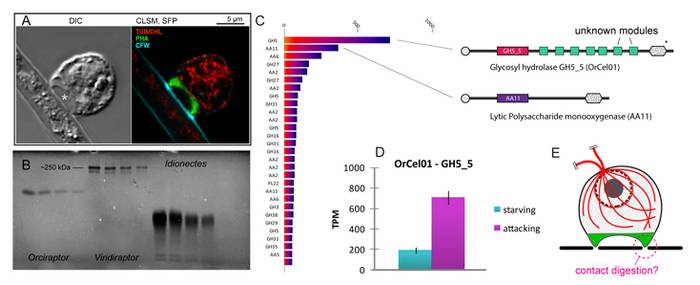
There is a diversity of parasitoid microbes that attack microalgae and fungi in aquatic and terrestrial ecosystems, forming an integral part of the microbial food web. As suggested by environmental sequencing, many of these highly-specialised interactions are still poorly known. This applies in particular to algivorous protoplast feeders, phagotrophic microeukaryotes that perforate the cell walls of algae and consume the cell contents. During the past years we made progress in understanding the phylogenetic diversity, cellular structure and ecology of protoplast feeders. However, it is still unclear how exactly protoplast feeders recognise their prey and perforate the prey cell wall. Transcriptomic data about the viridiraptorid amoeboflagellate Orciraptor agilis (Rhizaria) suggest that carbohydrate-active enzymes (CAZymes) mediate binding to the prey cell wall and cell wall dissolution. The protein sequences of these CAZymes represent an ideal basis for taking the next experimental steps, to establish a refined model of how protoplast feeders recognise and perforate the cell walls of their prey. Employing heterologous expression systems, we will produce recombinant candidate CAZymes and assay their activity on artificial and natural substrates. To establish the cellular localisation of these proteins in Orciraptor, antibodies will be raised and used for immunofluorescence microscopy. Furthermore, we will investigate the evolution of protoplast feeders by exploring the transcriptomes of the bacterivorous relatives of Orciraptor, specifically screening for cellulases and carbohydrate-binding proteins. Finally, we will expand our research to the vampyrellid amoebae to study how these versatile predatory amoebae adapted to diverse prey types. We will compare the feeding ecology and CAZyme expression in a wide phylogenetic framework and test with differential expression analyses if generalist vampyrellids regulate CAZyme expression in response to the prey cell wall biochemistry. By comparing the molecular toolkit of protoplast feeders of several distant phylogenetic lineages, we will finally understand how this fascinating feeding strategy evolved several times independently in the tree of life.
- Methods: Transcriptomics, cloning, protein biochemistry, enzymatic assays, immunocytochemistry, electron microscopy
- Relevant publications: Hess and Melkonian 2014, Busch and Hess 2017
Diversity and evolution of saccoderm desmids (Zygnematophyceae, Streptophyta)

The conjugating green algae (Zygnematophyceae) are the closest relatives of land plants and hence are of great evolutionary interest. Besides the popular placoderm desmids and the filamentous species, there is an underappreciated diversity of unicellular zygnematophytes with a much “simpler” morphology and smooth cell walls – traditionally referred to as “saccoderm desmids”. These saccoderm desmids have a broad geographic distribution and are ecologically diverse. Many species inhabit terrestrial habitats such as dead wood, rock surfaces and glacial ice. Furthermore, several of the saccoderm genera have turned out to be highly polyphyletic and are typically poorly captured by environmental sequencing approaches. With morphological and molecular methods, we shed some light on these inconspicuous yet important members of the algal flora and characterise diverse evolutionary lineages of structurally simple zygnematophytes.
- Methods: Light microscopy incl. fluorescence microscopy, molecular phylogenetics, taxonomy
- Relevant publications: Busch & Hess 2022a, 2022b
Cellular changes during the production of an extracellular sunscreen pigment found in terrestrial green algae
The ‘conjugating green algae’ (Zygnematophyceae) are the closest relatives of land plants and, hence, highly interesting from an evolutionary perspective. Given the immense structural and ecological diversity of these algae, their adaptations to the various habitats are still poorly understood. In a new genus of unicellular, terrestrial zygnematophytes (Serritaenia) we, recently, discovered an extracellular sunscreen pigment that can be experimentally induced by ultraviolet radiation. In the proposed project we will explore the cellular changes during the production of this yet unknown compound with comparative transcriptomics, metabolomics as well as electron microscopy. These techniques will enable us to (1) identify metabolic pathways involved in the production of Serritaenia’s sunscreen, (2) determine the compound itself (or fragments), (3) shed light on the molecular components involved in UV perception, and (4) gain deeper insights into the secretion of pigmented mucilage in Serritaenia. With this, we will uncover the cellular and molecular basis of a unique photoprotective strategy in the zygnematophytes, and enable exciting comparisons with algal and plant model systems..
Funded by the German Research Foundation, DFG; 2021-2024
- Methods: Light- and electron microscopy, differential expression analyses (transcriptomics), mass spectrometry
- Relevant publications: Busch & Hess 2021
The rotary eukaryotic flagellum of Idionectes vortex (Amoebozoa)
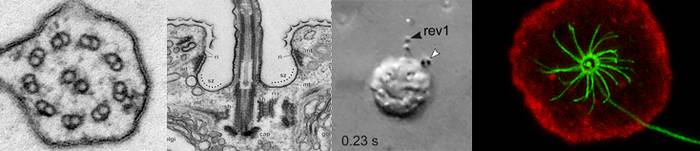
- Methods: Video microscopy, motion tracking experiments, transmission electron microscopy, immunocytochemistry, fluorescence microscopy
- Relevant publications: Hess et al. 2019
Protist-bacteria endosymbioses
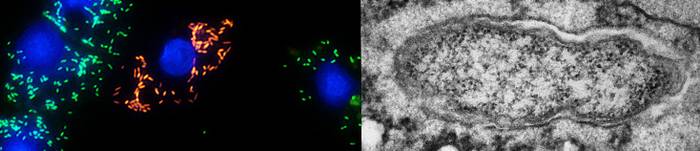
- Methods: Transmission electron microscopy, design of molecular probes, fluorescence in situ hybridisation (FISH), confocal laser scanning microscopy, molecular phylogenetics, taxonomy of prokaryotes
- Relevant publications: Hess et al. 2016, Hess 2017a, Muñoz-Gómez et al. 2019